Welcome to PBxplore’s documentation!¶
PBxplore is a suite of tools dedicated to Protein Block analysis.
Protein Blocks (PBs) are structural prototypes defined by de Brevern et al [1]. The 3-dimensional local structure of a protein backbone can be modelized as an 1-dimensional sequence of PBs. In principle, any conformation of any amino acid could be represented by one of the sixteen available Protein Blocks.
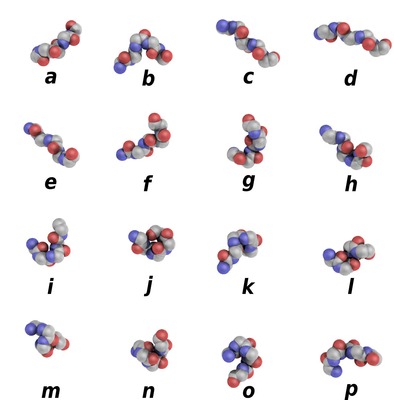
Schematic representation of the sixteen protein blocks, labeled from a to p (Creative commons 4.0 CC-BY).
PBxplore provides both a Python library and command-line tools (Utilization). For some features (reading trajectory, plots), PBxplore requires some optional dependencies (Installation). Basically, PBxplore can:
- assign PBs from either a PDB or either a molecular dynamics trajectory (PB assignation).
- use analysis tools to perform statistical analysis on PBs (Statistics).
- use analysis tools to study protein flexibility and deformability (Analyzis).
Contact & Support¶
PBxplore is a research software and has been developped by:
- Pierre Poulain, DSIMB, Ets Poulain, Pointe-Noire, Congo
- Jonathan Barnoud, University of Groningen, Groningen, The Netherlands
- Hubert Santuz, DSIMB, Paris, France
- Alexandre G. de Brevern, DSIMB, Paris, France
If you want to report a bug or request a feature, please use the GitHub issue system.
Licence¶
PBxplore is licensed under The MIT License.
| [1] |
|